Research
Parkinson’s disease
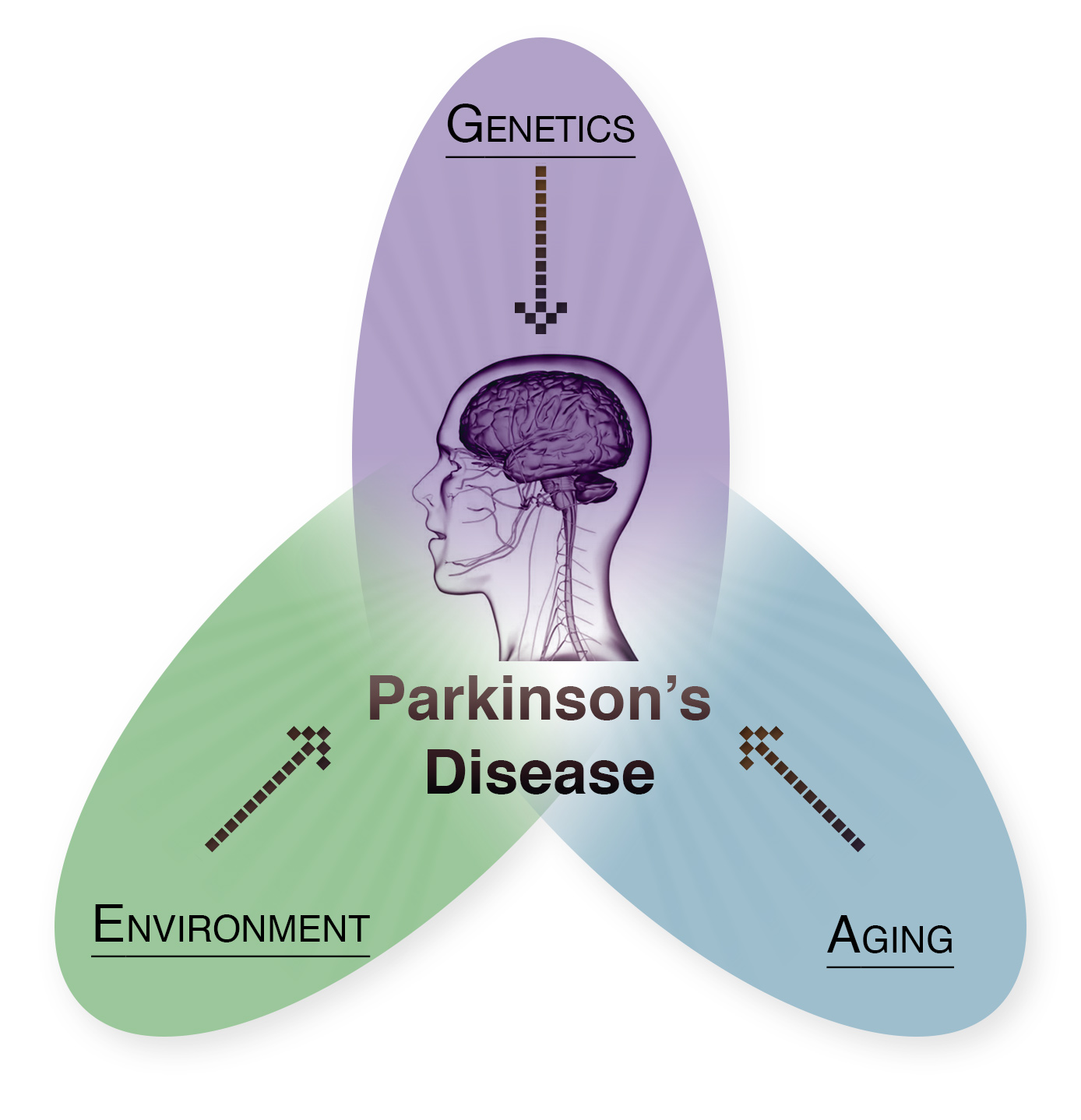
As the most common neurodegenerative movement disorder, Parkinson’s Disease (PD) represents a major clinical and societal burden, leading to progressive locomotor deficits, loss of dopaminergic neurons in the midbrain, and poor quality of life for the patients. The hallmark neuropathologic feature of this complex disease is the accumulation of α-synuclein (αSyn) protein aggregates in Lewy bodies in the PD brain. Rare SNCA point mutations and genomic multiplications are linked to familial PD cases with high penetrance and therefore constitute major genetic risk factors for PD. However, the preponderance of PD cases is apparently sporadic, suggesting the involvement of epigenomic factors for the development of PD and synucleinopathies. Moreover, the association of other environmental factors including aging, high fat diet and exercise, further underscores the importance for the consideration of epigenomic mechanisms in PD etiology and progression.
In addition to animal models, we used cell lines and cohorts from Parkinson's patients in a BMBF-ANR-CIHR project (decipherPD; 01KU1503A) to better understand the interplay of genes, environmental factors and aging in PD using transcriptome and epigenome sequencing. Using RNA-sequencing, we observed cell type-specific gene expression changes in neurons and glial cells in a BAC SNCA mouse model that were largely prevented by long-term environmental enrichment (Wassouf et al., 2018). Furthermore, we were able to show that high-fat diet leads to extensive adaptations of gene expression in the brain of healthy mice, but that this adaptation fails almost completely in a Parkinson's mouse model (Kilzheimer et al., 2023). In LUHMES cells differentiated into dopaminergic neurons, we were able to demonstrate genome-wide DNA methylation and hydroxymethylation changes associated with the overexpression of two Parkinson's-linked alpha-synuclein mutations (Schaffner et al., 2022).
The neuropathology of Parkinson's is based on alpha-synuclein protein aggregates in Lewy bodies. It is possible that the increased expression of the associated SNCA gene, controlled by DNA methylation, among other things, leads to the increased levels of alpha-synuclein. The central approach of a DFG-funded project in collaboration with Albert Jeltsch is to achieve repression of SNCA using epigenome editing in order to alleviate Parkinson's symptoms and restore cell and tissue homeostasis. For this purpose, DNA methylation and gene expression are profiled in animal and in cell models of PD in order to assess the effect of the EpiEditor and the therapeutic potential of this approach.
Huntington’s disease
In epiROM, a project in cooperation with Dr. Jonas Neher financed by the Baden-Württemberg Foundation, we investigated epigenetic reprogramming of microglia after immune stimulation by lipopolysaccharides and high-calorie diet in Parkinson's and Huntington's. In the context of Alzheimer's, the Neher group has already shown that peripherally applied inflammatory stimuli induce acute immune training and tolerance in the brain and lead to an epigenetic reprogramming of microglia that lasts for at least six months. In the current project, we examine comparable effects in Parkinson's and Huntington's rat models. Data have already been produced at bulk and single-cell level in order to be able to resolve disease-associated cell populations and transcriptional and epigenetic changes at the single-cell level.
Some cellular processes appear to be similarly disrupted in several neurodegenerative diseases, including Parkinson's and Huntington's disease. In addition, the influences of an enriched environment apparently produce comparable protective effects, which, however, are still largely unexplained at the molecular level. Therefore, in collaboration with Prof. Huu Phuc Nguyen (Ruhr University Bochum), we are investigating whether the key genes identified in Parkinson's also assume protective functions in Huntington's and to what extent they can phenotypically counteract the progression of the disease (Novati et al., 2018). We measure DNA methylation changes parallel to transcriptome changes in different brain regions of Parkinson's and Huntington's disease rat models and integrate these data to a cross-disease understanding.
Ageing
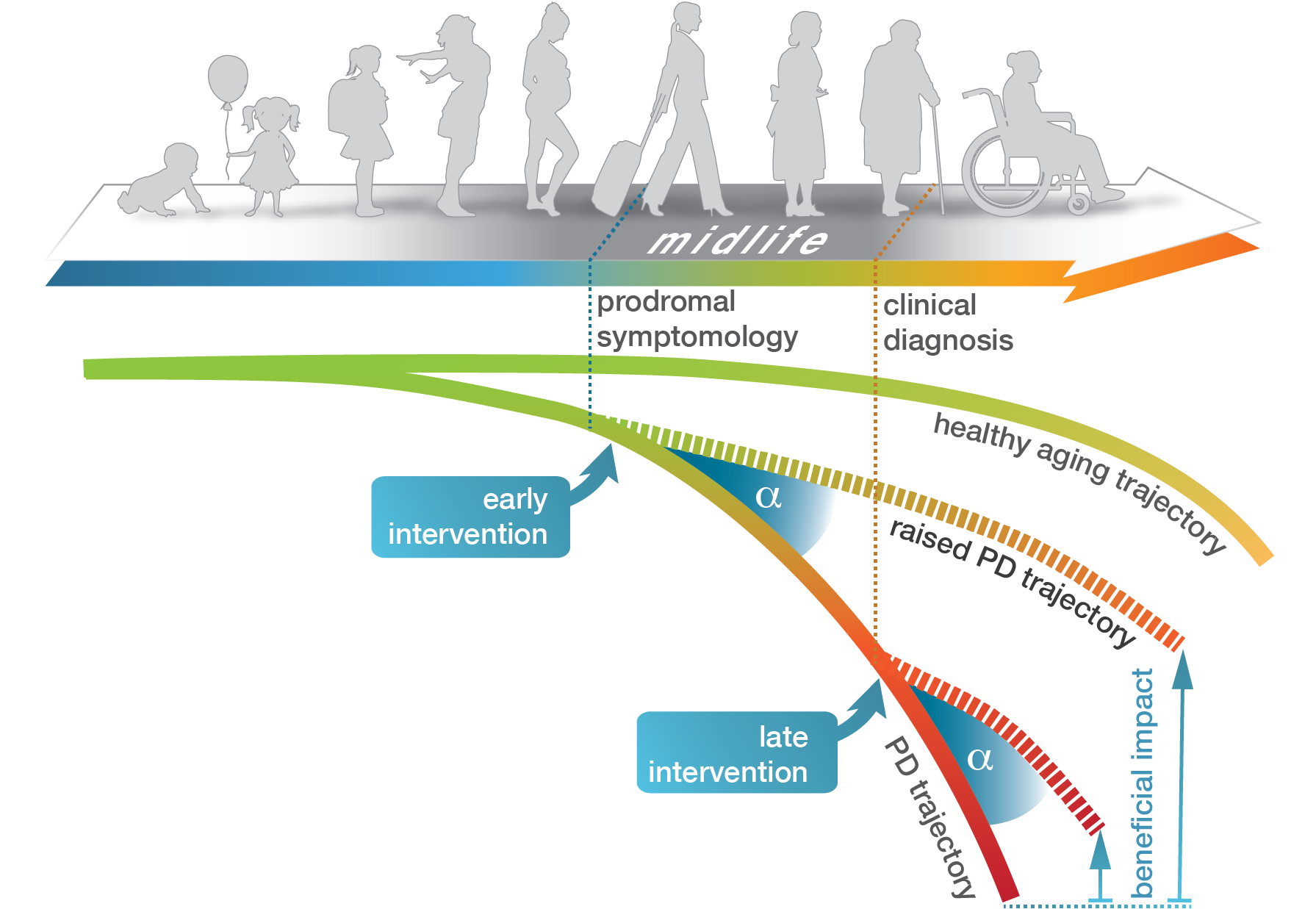
In addition to environmental influences, aging processes play an important role in the progression of Parkinson's disease. In this context, we investigate the necessary adaptations at the epigenome and transcriptome level during healthy aging and determine molecular abnormalities that underlie the development of Parkinson's disease. Transcriptome changes in animal models indicate two characteristic forms of age-dependent gene expression changes in the context of Parkinson's disease: First, cellular processes seemingly activated too early that reflected advanced stages of age and, second, typical longitudinal adaptations of the system that no longer occurred during midlife (Hentrich et al., 2018). In a current DFG grant, we examine the essential adaptations at the epigenome and transcriptome level during healthy aging and determine molecular abnormalities that underlie the development of Parkinson's disease. We use samples from a Parkinson's rat model and post-mortem brain samples from Parkinson's patients.
Epigenetics in rare diseases
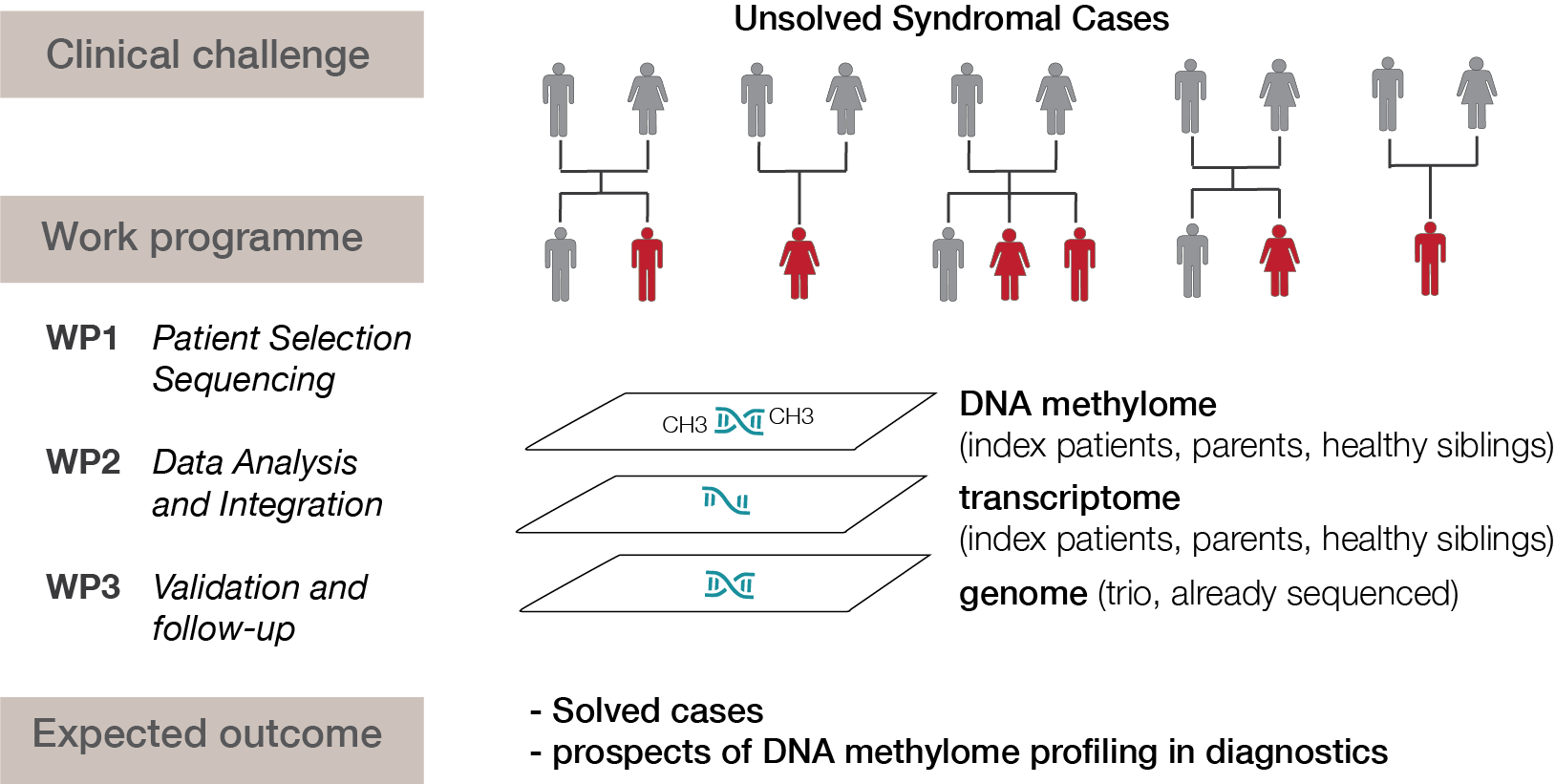
Although exome and genome sequencing are crucial in routine diagnostics to elucidate new disease genes and molecular diagnoses of rare diseases, they can only solve around 40% of all cases. The sequencing of other molecular layers - especially the epigenome - which is not yet part of routine diagnostics, could help to close this gap. Alterations in DNA methylation have long been known to be implicated in certain groups of constitutional syndromes such as imprinting disorders. Recently, it has been shown that other cases may result from DNA methylation defects at single loci (epi-variants) or exhibit syndrome-specific DNA methylation changes across multiple loci (epi-signatures). This approach could help understanding rare diseases and is currently being used in the solveRD consortium (solving the unsolved rare diseases, solve-rd.eu). In this consortium, we lead the epigenomics workgroup that evaluates DNA methylation in unsolvable syndromes.
In addition, in a DFG project together with Dr. Rebecca Buchert-Lo, we chart the DNA methylome in unsolved syndromal cases and evaluate how epigenetic profiling can be integrated into clinical diagnostics of rare diseases.